Multiple mechanisms including oxidative stress, inflammation and excitotoxicity contribute to brain injury during ischemic stroke [1,2]. Regardless of the specific mechanism at play, the ultimate fate of most affected brain cells is death, unless timely and appropriate interventions are implemented. Recently, studies from other labs and ours have identified various forms of cell death, such as apoptosis, necroptosis, pyroptosis, and ferroptosis, occurring in the brain during ischemic stroke [[3], [4], [5], [6], [7], [8]]. Each of these forms of cell death plays a pivotal role in the pathophysiology of ischemic stroke.
The cell death pathways implicated in ischemic stroke are highly interconnected, functioning within a complex network with extensive crosstalk. Key proteins involved in the pathway crosstalk, such as receptor-interacting protein kinase 1 (RIPK1), Caspases, and Bcl-2 family proteins, are involved in regulating multiple cell death pathways. For instance, RIPK1 is a key mediator of necroptosis but also modulates apoptosis and pyroptosis [9,10]. Caspase-8 is classically considered an apoptotic Caspase but also cleaves and activates gasdermin D, a key mediator of pyroptosis [11]. Similarly, Caspase-3 has roles in both apoptosis and pyroptosis [10,11]. This intricate network with extensive crosstalk between regulators complicates the development of targeted therapies, as the consequences of inhibiting a single node within the network are not fully understood. Thus, elucidating the interplay between diverse cell death pathways and their key regulatory proteins is imperative for identifying therapeutic targets that can potentially modulate cell death in many forms of cell death in stroke, and thus develop effective stroke treatments.
The traditional drug discovery process, often taking over a decade, struggles to meet the need for rapid development of effective treatments [12]. To accelerate this process, the field is increasingly leveraging computational methodologies. Traditional computational techniques, such as molecular docking simulations, utilize 3D structures of drug-target pairs to identify binding sites. However, these methods require known high-resolution 3D structures of both targets and drugs, which may not always be available [13]. An alternative strategy involves extracting features from drugs and targets to predict potential interactions. Drug-target interaction (DTI) prediction methods have evolved from Support Vector Machine (SVM) and Random Forest (RF) algorithms to more recent deep learning techniques [14,15]. These methods treat DTI prediction as either a binary classification problem, where a drug candidate can or cannot bind to a target, or as a regression task, where the dissociation constant is used as the binding affinity for prediction. The field of DTI prediction has seen significant advancements in recent years, with a plethora of deep learning-based methodologies emerging. These range from models based on deep belief networks, such as DeepDTI, to those leveraging graph-based and knowledge graph-based recommendation systems [[16], [17], [18]]. Additionally, end-to-end deep collaborative learning models [19], capsule network-based approaches [20], multimodal methods [21], and attention-based networks have also been developed [22], broadening the scope and potential of DTI predictions. However, the main challenge in applying these methods to stroke drug discovery is the complexity of selecting appropriate models and validating their predictions because of the huge number of potential drug-target interactions and the difficulty of evaluating model performance in the real world [23]. From a biological standpoint, the differences in performance among various models are often minor, yet these slight variations can lead to significant differences in their predictions.
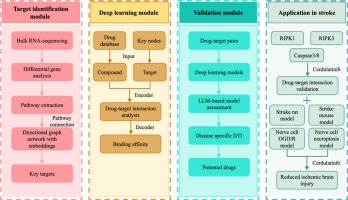
To address stroke drug discovery challenges, we introduce strokeDTI, a novel hybrid framework that integrates multiple deep learning architectures with transcriptomic data. It uses transcriptomic data from stroke models to extract activated pathways, forming a network of differentially expressed genes. The most common pathway combinations indicate key network bottlenecks, which are selected as potential targets. A unique feature of strokeDTI is its ability to predict stroke-specific drug-target interactions via incorporating multiple models and assessing their predictive stability in different cross-validation folds. Additionally, we use two validation modules to evaluate the predictive accuracy of each model in identifying effective interactions. Using this integrated approach, we identified Cerdulatinib as a potential anti-stroke agent, which was validated in mouse and rat ischemic stroke models. These findings highlight strokeDTI’s potential for disease-specific drug-target identification.